Abstract
Background: Most CLL patients do not need treatment at the time of diagnosis; however a group of CLL patients progress rapidly and respond poorly to conventional CLL treatments (Parikh, Semin Oncol, 2016) . Mutation in IGHV genes (mutated CLL) is a predictor for better disease outcomes compared with unmutated IGHV genes (unmutated CLL (Hamblin, Blood, 1999)). Targeted next-generation sequencing (Targeted NGS) has proven to be an accurate and reproducible technique when validated against the gold standard Sanger sequencing (Lesley-Ann Sutton, Haematologica, 2014). Both CD38 "a surface molecule" and CD49d "an alpha-chain of the integrin heterodimer VLA-4" are involved in CLL cell migration, adhesion and homing to secondary lymphoid organs. CD38 and CD49d are poor prognostic factors in CLL (Gentile, Br J Haematol, 2005; Gattei, Blood, 2008). There is physical and functional association between CD49d and CD38 expression (Zucchetto, Leukemia, 2012)
Aim: We aim to investigate the impact of IGHV status, CD49d and CD38 expression on treatment free survival (TFS) and time to first treatment (TTFT) in CLL.
Methods: We enrolled 23 patients with a confirmed diagnosis of CLL (WHO criteria). CLL cells were extracted from whole blood by negative selection using RosetteSep Human B Cell Enrichment Cocktail. Immunophenotyping was performed using Beckman Coulter FC500 Flowcytometer. CLL cells were identified by co- expression of CD5 and CD19. We checked the expression of CD38 and CD49d and we used a cut off of 7% or more for CD38 positivity and 30% or more for CD49d positivity as per previous studies (Gentile, Br J Haematol, 2005; Gattei, Blood, 2008). Targeted NGS was performed and reported using the ERIC (European Research Initiative on CLL) panel that includes: ATM, BIRC3, MYD88, NOTCH1, SF3B1, TP53, EGR2, POT1, NFKBIE and XPO1 [www.ericll.org]. IGHV mutational analysis was performed and patients with no or limited somatic hyper mutation burden (germline identity (GI) ≥ 98%) constituted "unmutated CLL" (U-CLL), whereas patients with GI < 98% constituted "mutated CLL", M-CLL).
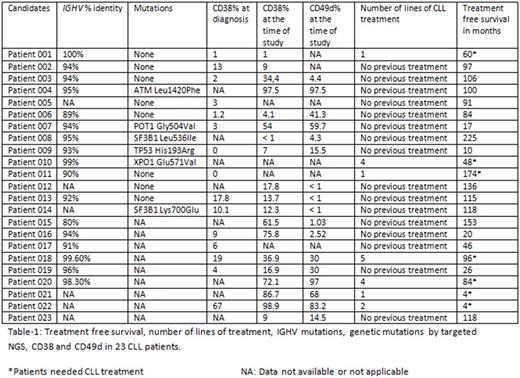
Results: Sixteen patients were treatment naïve while 7 patients received one or more lines of CLL treatment (Table-1). Treatment naïve patients had variable time since diagnosis that ranged between 17 and 225 months (Median: 91 months) and half of them had not required CLL treatment for more than 10 years. Our data showed that CD38 expression is dynamic and can increase, decrease or stay the same over the disease course (Table-1). We found that the level of CD38 was significantly higher in patients who required CLL treatment compared to patients who did not require CLL treatment (p=0.031) There was also a significant increase in CD 49d expression in patients who required CLL treatment compared to patients who did not require CLL treatment (p=0.023). All patients who needed CLL treatment and tested for both CD38 and CD49d (N: 4) showed strong double expression of both antigens. In stark contrast, only 15% (2/13) of the patients who never required CLL treatment showed double expression of both CD38 and CD49d. IGHV was unmutated in 23.5% (4/17) of patients. All the patients with unmutated IGHV required one or more lines of CLL treatment, while only 7.6% (1/13) patients with mutated IGHV required CLL treatment. Targeted NGS showed that 42.8% (6/14) of the patients carried a mutation (Table-1). We did not find any correlation between genetic mutations by NGS and treatment free survival (TFS). Only one patient with a mutation (XPO1) needed CLL treatment. The remaining 5 patients with mutations including high risk mutations like TP53 and ATM have so far not required treatment. None of the patients with both mutated IGHV and double negative CD38/CD49d required CLL treatment.
Conclusion: CLL patients with mutated IGHV and double negative CD38/CD49d have a good prognosis and long treatment free survival regardless of their genetic mutations. We suggest that combining IGHV mutational analysis with high risk immunophentotypic markers like CD38 and CD49d may offer an enhanced prognostic tool beyond that offered by genetic mutations in isolation in predicting treatment free survival (TFS) and time to first treatment (TTFT).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal